Miranda Kadwell(1), Matilde Fernandez(2), Helen F. Stanley(1), Ricardo Baldi(3),
Jane C. Wheeler(4,2)*, Raul Rosadio(2) and Michael W. Bruford(5,1)**
(1) Institute of Zoology, Regent’s Park, London NW1 4RY, UK
(2) Facultad de Medicina Veterinaria, Universidad Nacional Mayor de San Marcos, Apartado 41- 0068, Lima 41, Peru
(3) Centro Nacional Patagonico, Consejo Nacional de Investigacions Cienti¢cas yTechnologicas, 9120 Puerto Madryn, Argentina
(4) CONOPA, Los Cerezos 106, Salamanca-Lima 3, Peru
(5) Biodiversity and Ecological Processes Group, Cardi¡ School of Biosciences, Cathays Park, Cardi¡ CF10 3TL, UK
* Author for correspondence for camelids (webmaster@conopa.org).
** Author for correspondence for genetic analysis
(brufordmw@cardi¡.ac.uk).
The origins of South America’s domestic alpaca and llama remain controversial due to hybridization,
near extirpation during the Spanish conquest and di¤culties in archaeological interpretation. Traditionally,
the ancestry of both forms is attributed to the guanaco, while the vicu·a is assumed never to have
been domesticated. Recent research has, however, linked the alpaca to the vicu·a, dating domestication
to 6000^7000 years before present in the Peruvian Andes. Here, we examine in detail the genetic relationships
between the South American camelids in order to determine the origins of the domestic forms,
using mitochondrial (mt) and microsatellite DNA. MtDNA analysis places 80% of llama and alpaca
sequences in the guanaco lineage, with those possessing vicu·a mtDNA being nearly all alpaca or
alpaca^vicu·a hybrids. We also examined four microsatellites in wild known-provenance vicuña and
guanaco, including two loci with non-overlapping allele size ranges in the wild species. In contrast to the
mtDNA, these markers show high genetic similarity between alpaca and vicu·a, and between llama and
guanaco, although bidirectional hybridization is also revealed. Finally, combined marker analysis on a
subset of samples con¢rms the microsatellite interpretation and suggests that the alpaca is descended from
the vicu·a, and should be reclassi¢ed as Vicugna pacos. This result has major implications for the future
management of wild and domestic camelids in South America.
Keywords: domestication; alpaca; guanaco; llama; vicuña; mitochondrial DNA
1. INTRODUCTION
Four South American camelids are recognized today, two
of which are wild species, the guanaco (Lama guanicoe,
MÏller 1776) and the vicuña (Vicugna vicugna, Molina
1782), and two of which are domestic forms, the alpaca
(Lama pacos L.) and the llama (Lama glama L.), whose
evolutionary origins are debated (Wheeler 1995). The
guanaco and vicuña diverged from a common ancestor
around two million years ago, and are the only representatives
of the lamini to survive the Pleistocene period
(Stanley et al. 1994; Wheeler 1995).
Archaeozoological
evidence from the central Peruvian Andes links alpaca
origins to the vicuña at 6000^7000 years before present
(Wheeler 1995). Because all potential ancestral forms are
extant, South American camelid domestication represents
an unusual and useful opportunity to gain insight into the
origin and biodiversity of domesticated animals, an issue
which is of increasing interest due to the recognized
potential economic bene¢ts of indigenous genetic
resources and the threats that face marginal and extensive
agriculture today (Hall & Bradley 1995). In contrast with
many other domestic farm animals, there is no written
history associated with the llama and alpaca.
Orally
transmitted herding knowledge was largely lost during
the Spanish conquest, and breeds disappeared as both the human and the native domestic live stock populations
were reduced by 80^90% during the ¢rst 100 years of
contact (Wheeler 1995). At present, although llama and
alpaca rearing is a central element of the economy in the
high Andes, it is often not pro¢table due to the poor
quality of the animals and their fibre. The reconstruction
of fine-fibre breeds and the breeding strategies needed are
therefore uniquely dependent upon the contributions of
archaeozoology and genetic analysis (Wheeler et al. 1995).
Understanding the domestication of the llama and
alpaca using morphological analyses has been hampered
by the lack of species-specifc post-cranial skeletal characteristics,
and because archaeological remains are often in
poor condition (Bahn 1994). Fibre analysis has been
pursued at sites where £eece has been preserved
(Dransart 1991a; Reigadas 1993; Wheeler et al. 1995), but
the decimation of traditional llama and alpaca breeds at
the time of the conquest, and probable subsequent hybridization
among the survivors, make inference from
phenotypic characters problematic.
Molecular genetic analysis using methods such as mitochondrial
DNA (mtDNA) sequencing and microsatellite
and Y-chromosome analysis has recently proved illuminating
when examining the origins of modern domestic
livestock, even in the face of often overwhelming recent
changes in domestic populations due to genetic drift,
selection and/or hybridization (e.g. Bradley et al. 1996;
MacHugh et al. 1997; Hiendleder et al. 1998; Luikart et al. 2001; Vila et al. 2001). Here, we apply mitochondrial
cytochrome b sequence and microsatellite typing to the question of the evolutionary relationships between and
domestication of South American camelids throughout
their range in South America today.
2. MATERIAL AND METHODS
Sample collection sites span as far as possible the geographical
range of the two wild species (see ¢gure 1). Our
sample comprises: two vicu·a subspecies (V. v. vicugna and
V. v. mensalis) from Argentina, Chile and Peru, guanaco
(L. g. guanicoe and L. g. cacsiliensis) from Argentina and Peru,
llama (a range of morphological types) from Argentina, Bolivia
and Peru, alpaca (including `suri’ and `huacaya’ £eece types)
from Argentina, Bolivia, Chile and Peru, and 10 known hybrids
(seven between llama and alpaca (locally known as wari) and
three between alpaca and vicu·a (locally known as pacovicu·a). Samples were taken from only those individuals whose
phenotype conformed to accepted morphological criteria for
domestic forms. The localities and genetic data for each individual
included in this study can be found at http://www.cardi¡.
ac.uk/biosi/research/biodiversity/sta¡/mb.html.
(b) MtDNA analysis
The phylogenetic afiliations of the llama and alpaca were¢rst confirmed by sequencing a short (158 bp) but highly informative
region of the cytochrome b gene of the mitochondrial
genome sensu Stanley et al. (1994) (GenBank accession numbers
U06425^U06430). In total, 211 South American camelids were
analysed from throughout the geographical range (comprising
21 guanaco, 42 vicuña, 54 llama, 84 alpaca and the 10 hybrids
described in table 1). One Arabian camel was also analysed.
Briefly, DNA was extracted from blood or skin using standard
proteinase-K digestion followed by organic extraction using
phenol and phenol^chloroform, and total DNA was precipitated
in 100% ethanol (Stanley et al. 1994; Bruford et al. 1998). DNA
samples were stored in TE bu¡er (10mM Tris-HCl, 1mM
EDTA, pH 8.0). The cytochrome b primers L14724 and H14900
were used for polymerase chain reaction (PCR), which was
carried out as in Stanley et al. (1994). PCR products were purified and DNA sequencing was carried out as previously
described (Stanley et al. 1994).
Sequences were aligned manually, and unique haplotypes
have been deposited in GenBank under accession numbers
AF373809^AF373833. Mitochondrial haplotype divergence and
frequencies were analysed for the guanaco, vicu·a, llama and
alpaca samples as previously described (Stanley et al. 1994), and a
minimum-spanning network (Kruskal 1956; Bandelt at al. 1999)
was generated using the program MINSPNET (Exco¤er 1993;¢gure 2). The distribution patterns of domestic South American
camelid haplotypes were then compared with those of the wild
South American camelid sample.
(c) Microsatellite analysis
Since the strict maternal inheritance of mtDNA in most
mammals restricts its use in studies of hybridization, especially
in domestic livestock (e.g. MacHugh et al. 1997), we also applied
nuclear DNA markers. Four microsatellite loci (YWLL 38,
YWLL 43,YWLL 46 and LCA 19; Lang et al. 1996; Penedo et al.
1998) were typed for 669-771 individuals, including the 211
individuals for cytochrome b (table 1).
The genetic distances
between the four South American camelid taxa were measured
in three ways: first, by using 1-(proportion of shared alleles); second, by using Reynold’s distance (Reynolds et al. 1983), a
measure commonly used in livestock analysis where genetic drift
has a major impact on allele frequencies; and finally, by using δμ2 (Goldstein et al. 1995), which calculates population subdivision
using allele size di¡erences under a stepwise mutation
model. All distances were estimated using the program
MICROSAT v. 1.5d (Minch 1999).
Factorial correspondence analysis was then performed on
pairwise allele frequency di¡erences using GENETIX v. 4 (Belkhir
1999). Here, the genetic di¡erentation between populations (in
this case vicu·a, guanaco, llama and alpaca) is expressed as
factors, which explain the correspondence between samples in a
number of dimensions (Benzécri 1973).
Thus, the relationships
among populations can be judged by examining how individuals
from each population cluster in two or three dimensions.
(d) Combined analysis
To assess the extent of introgression in llama and alpaca
populations, we used both the microsatellite and the mitochondrial
data to calculate the admixture proportions mC and mY
(mC: estimator based on allele frequencies, Chakraborty et al.
(1992); mY: estimator based on the number of substitutions for
mtDNA and squared allele size di¡erence for microsatellite
data, Bertorelle & Exco¤er (1998)) for alpaca and llama compared
with the vicu·a and guanaco. This was implemented
using the program ADMIX 1__0 (Bertorelle 1998).
Finally, to examine the concordance between mitochondrial
and microsatellite data and patterns of introgression in more
detail, we returned to the 211 individuals typed for both mtDNA
and microsatellites. Genotypes were coded `V’ (vicuña) or `G’
(guanaco) for mtDNA and `V’, `G’ or `H’ (hybrid) for YWLL 46
and LCA 19, depending on their allele sizes with reference to
the guanaco and vicuña ranges, and we examined the data for
each locus separately and combined.
3. RESULTS
(a) Mt DNA
We found 26 unique haplotypes within the 211 South
American camelids analysed from throughout the geographical
range. Uncorrected distances within the South
American camelids ranged from 0.006 (one substitution)
to 0.089 (14 substitutions).
The minimum-spanning
network (figure 2) recovers two groups, recapitulating
the reciprocally monophyletic clades found previously
(Stanley et al. 1994). The first group contains all vicuña
(V), and the other contains all guanaco (G). Both groups
are connected by 21 substitutions to the Arabian camel.
The domestic South American camelids are found in
both groups, but 81% (120 out of 148, including 61 out of
84 alpaca) are found within the `G’ group. A minority
(28 out of 148) comprising alpaca (n=23), pacovicuña
(n= 3) and llama (n = 2) are found within the `V’ group.
(b) Microsatellites
A feature of the microsatellite loci analysed here is the
large number of private alleles found when comparing
vicu·a and guanaco. These private alleles, which range
between 33% for YWLL 38 and 100% for LCA 19 and
YWLL 46, occupy predominantly di¡erent allele size
ranges. Figure 3a(i) and 3b(i) show allele frequency
histograms for LCA 19 and YWLL 46 for wild vicuña
and guanaco: the allele sizes do not overlap between the
(a)
 
(b)
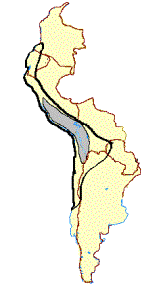 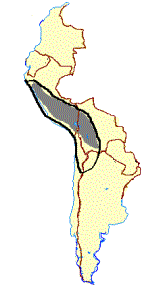
 
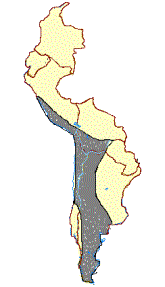
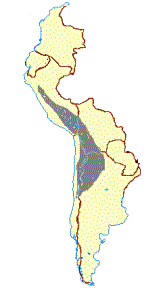
Figure 1. (a)Geographical distribution and photographs of the guanaco (L. g. cacsilensis) (left) and vicu·a (V. v. mensalis) (right). Map
taken fromWheeler et al. (1995). Photographs:guanaco, P. Daulesberg; vicu·a, J.C.W. Shaded areas mark the extent of present
ranges, solid circles represent Pleistocene and Early Holocene fossil localities. (b)Geographical distribution and photographs of the
llama (left) and alpaca (right). Map taken fromWheeler et al. (1995). Photographs: J.C.W. Solid linesmark the distribution prior to
1532, dashed linesmark the present range, solid circles represent archaeological sites containing llama and alpaca remains.
two species. These loci therefore provide potentially
powerful tools for the discrimination of ancestral
genomes in modern domestic stock. Figure 3a(ii) and 3b(ii) show histograms for the same loci in llama and
alpaca, which display similar patterns. However, the
patterns of genetic similarity are in contrast to those
Table 1. South American camelid samples analysed for mitochondrial DNA (cytochrome b) and four microsatellite
loci.
 |
| |
cytocromeb mtDNA |
YWLL 38 |
LCA a |
YWLL 46 a |
YWLL 43 |
 |
| vicuña |
42 |
434 |
439 |
438 |
440 |
| Guanaco |
21 |
112 |
120 |
122 |
115 |
| wari |
7 |
7 |
7 |
7 |
7 |
| pacovicuña |
3 |
3 |
3 |
3 |
3 |
| llama |
54 |
50 |
60 |
60 |
60 |
| alpaca |
84 |
63 |
138 |
141 |
103 |
 |
a Indicates loci with non-overlapping allele size ranges.
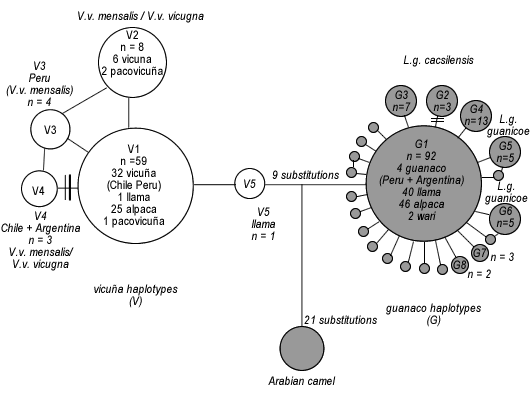
Figure 2. Minimum-spanning network representing the relationships between cytochrome b mitochondrial haplotypes as circular
nodes, where the number of substitutions (if greater than one) are represented by multiple dashes or numbers on each connecting
line. The relative frequency of each haplotype is represented by the area of the circle. Unfilled circles indicate vicuña haplotypes,
and filled circles represent guanaco haplotypes. Wild samples are specifically referred to where present. Phylogenetic analysis
(maximum parsimony and neighbour joining based on uncorrected p, JC and K2P sequence distances) recovered an equivalent
pattern to the network, and high (490%) bootstrap support was always found for the major split between `V’ and `G’ haplotypes
(not shown).
revealed by mtDNA. Visual inspection reveals strong
similarities between the allele size distributions of vicuña
and alpaca, and between guanaco and llama. For
example, for YWLL 46, the 98 bp allele has a frequency
of 0.95 in the vicuña sample and 0.75 in the alpaca,
while the 104 and 106 bp alleles have a combined
frequency of 0.91 in the guanaco and 0.64 in the llama.
Analysis of all four loci reveals that genetic distances
between the vicu·a and alpaca and between the
guanaco and llama (table 2) are almost always much
lower than those between the vicuña and guanaco, the
vicu·a and llama or the guanaco and alpaca. Distances
between the alpaca and llama are intermediate, with the exception of δμ2, where the distance between the
guanaco and llama is slightly larger.
However, a second feature of the histogram in figure 3b
is the presence, at low frequencies, of `vicuña’ alleles in
the llama sample and of `guanaco’ alleles in the alpaca
sample. For example, in LCA 19, the 99 bp and 103 bp
`vicu·a’ alleles are present at a combined frequency of
0.063 in the llama sample, and the 87 bp `guanaco’ allele
has a frequency of 0.119 in the alpaca. The presence of
these alleles in our sample con¢rms bidirectional introgression
in both domestic forms.
A striking pattern emerges from the factorial correspondence
analysis (¢gure 4), where almost half of the
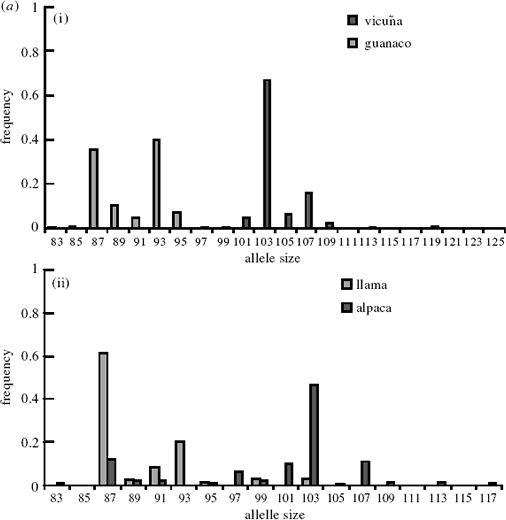
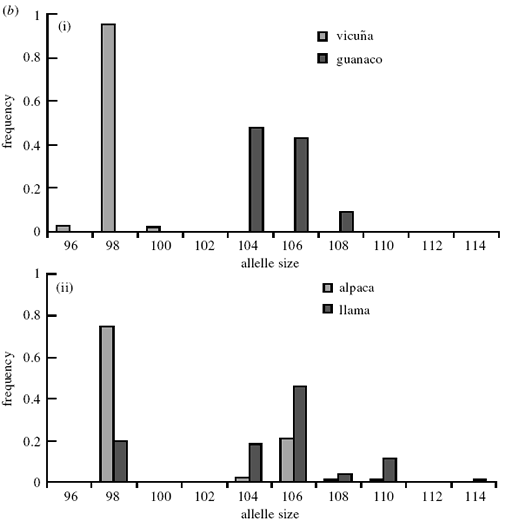
Figure 3. Allele frequency histograms for a large sample of (i) vicuña and guanaco and (ii) llama and alpaca for (a) LCA 19 and
(b) YWLL 46. Distributions were generated using the following: guanaco, n = 104 (LCA 19) and n = 177 (YWLL 46); llama,
n = 56 (LCA 19) and n = 58 (YWLL 46); alpaca, n = 80 (LCA 19) and n = 82 (YWLL 46); and vicuña, n = 227 (LCA 19) and
n = 231 (YWLL 46).
Table 2. Pairwise genetic distances between the four South American camelids. (a) Above diagonal: Reynolds
distance; below diagonal: δμ2. (b) Below diagonal: 17proportion of shared alleles.
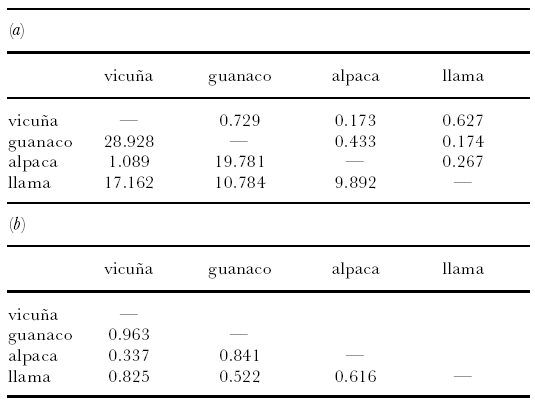
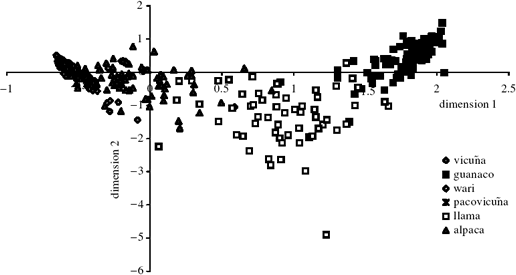
Figure 4. Two-dimensional factorial correspondence plot for allele frequencies at four microsatellite loci in all South American
camelids. Almost half of the explained correspondence (15%) is found in factors 1 and 2, represented on the horizontal and
vertical axes, respectively.
explained variance (15%) is found in factors 1 and 2,
represented on the horizontal and vertical axes, respectively.
It can be clearly seen that the guanaco (black
squares) and vicuña (grey, ¢lled diamonds) form two
tightly clustered and highly distinct groups. The alpaca
(grey, filled triangles) also form a cohesive group and
cluster strongly with the vicuña. In contrast, the llamas
(open squares) and hybrids form a much more di¡use
group. The llamas, although tending to cluster with
guanaco on axis 1, are more intermediate with respect to
the wild species when compared with the alpaca sample,
and are also the most genetically di¡use group on axis 2.
(c) Combined data
The admixture results are non-concordant between the
mitochondrial and microsatellite analyses in the alpaca,
where the estimated microsatellite proportion of vicuña genome is two to three times higher than the proportion
estimated using mtDNA (0.310 ± 0.121 for mtDNA;
0.903 ± 0.108 for mC and 0.823 ± 0.087 for mY for microsatellites).
Furthermore, in the llama, although both estimates
are relatively low, the microsatellite admixture
proportions are an order of magnitude higher
(0.0231 ± 0.242 for mtDNA; 0.220 ± 0.088 for mC and
0.389 ± 0.109 for mY for microsatellites). Importantly, it is
evident using both mC and mY that the proportion of
vicuña DNA is much lower in llama than in alpaca.
The combined three-locus analysis of the 211 individuals
produces striking results (table 3). Out of the 54
llamas, 52 (96%) possessed a `G’ mtDNA haplotype, and
47 (90%) and 33 (61%) possessed a pure `G’ genotype for
LCA 19 and YWLL 46, respectively. Out of the 84
alpacas, only 23 (27%) possessed a `V’ mtDNA haplotype,
while 63 (75%) and 66 (79%) possessed pure `V’
Table 3. Three-locus genotypes for samples where all three types of data are available. Loci are ordered
mitochondrial DNA, LCA 19 and YWLL 46; so, for example, GVH indicates guanaco mitochondrial DNA, vicuña
genotype at LCA 19 and a hybrid genotype at YWLL 46 (X signi¢es that the sample could not be typed).
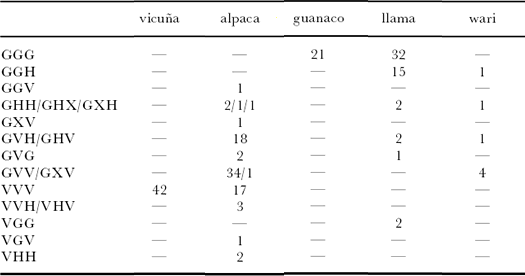
LCA 19 and YWLL 46 genotypes, respectively. Out of
the llamas tested in this study, 32 (59%) exhibited a
`GGG’ three-locus genotype; only 17 (20%) alpacas
exhibited a`VVV’ three-locus genotype. Extensive nuclear
introgression is detected in the llamas studied here, with
20 (37%) showing one or more `vicu·a’ alleles at LCA 19
and/or YWLL 46. In contrast, much of the presumed
introgression in the alpaca is mitochondrial, with 34
(40%) samples showing a `GVV’ three-locus genotype.
4. DISCUSSION
The data resulting from this study present a complex
picture of how modern-day domestic South American
camelid genetic diversity has been shaped by domestication,
selection, hybridization and genetic drift, with the
added uncertain impact of events relating to the Spanish
conquest. Such complexity is not without precedence in
studies of domestic livestock (e.g. MacHugh et al. 1997),
and requires detailed analysis and cautious interpretation.
Taken in isolation, the expanded mtDNA analysis
presented here largely supports the findings of Stanley et
al. (1994). Wild vicu·a and guanaco mtDNA is reciprocally
monophyletic with 5.8-8.9% uncorrected sequence
divergence being found between the two lineages, recapitulating
the suggestion in Stanley et al. (1994) that these
species diverged from a common ancestor two to three
million years ago. Furthermore, the ¢nding that nearly
all modern llamas possess a `guanaco’ haplotype is also
supported by the present dataset (table 3), where all
except two llamas from a sample of 54 individuals had
guanaco mtDNA. However, our much expanded alpaca
dataset reveals a di¡erent pattern from that previously
found, with only 27% of individuals possessing vicuña
mtDNA (table 3), in contrast to the 50% reported by
Stanley et al. (1994). It is worth noting here that our
sample comprises 84 individuals (as opposed to 14)
sampled from throughout the geographical range.
In
isolation, the ¢nding that a large proportion of modernday
alpacas possess guanaco mtDNA is in accordance
with hypotheses that alpacas, in common with llamas,
are descended from the guanaco. However, clearly, as in
Stanley et al. (1994), the presence of substantial numbers of alpaca possessing vicuña mtDNA also raises the possibility
that the alpaca is of mixed origin or has undergone
substantial hybridization during domestication or subsequently.
However, the limitations of mtDNA, which is
maternally inherited, in the context of gene flow and
evolution in domestic populations are obvious, since
historical and modern-day agricultural practices have
often used phenotypically desirable males to sire large
numbers of females. Therefore, analysis of biparentally
inherited markers (in this case microsatellites) capable of
discriminating between the wild ancestors was desirable.
The microsatellite data provide a contrast to the
mtDNA data, and the existence of two loci with nonoverlapping
allele size ranges in the wild ancestors
allowed us to compare patterns of divergence in relatively
large numbers of domestic animals. In contrast to the
mtDNA results, visual inspection of allele frequency
distributions, genetic-distance analysis using three
measures with contrasting models and assumptions, and
factorial correspondence analysis all reveal a striking
similarity between the alpaca and the vicuña (figures
3a,b, 4 and table 2a,b).
Each genetic-distance estimate is
lowest for the alpaca-vicuña comparison, and the
factorial correspondence analysis (figure 4) shows that the
alpaca and vicuña occupy an almost overlapping region
of the plot. These data, therefore, point towards a very
close genetic afinity between the alpaca and vicuña, a¢nding in complete contrast to the mtDNA data.
The microsatellite data in part agree with the mtDNA
data by supporting a close relationship between the llama
and guanaco. Out of the genetic-distance estimates, both
Reynold’s and allele sharing distances are second lowest
for the guanaco-llama comparison (table 2a,b), and the
Reynold’s distance estimate is almost identical to that for
the alpaca-vicuña comparison. However, other data are
more equivocal, with the factorial correspondence plot
revealing a dispersed pattern for the llama, which could
be described as intermediate between the vicuña and
guanaco, and the δμ2 distances being slightly lower for
the llama-alpaca comparison than for the llama-guanaco comparison. Although none of the above are
indicative of a close relationship between the llama and
vicuña, they suggest either that there has been nuclear gene flow between the llama and vicuña (or, more likely,
between the llama and alpaca) or that the ancestral stock
for the present-day llama was genetically highly diverse
and that this is re£ected in present-day microsatellite
allelic diversity.
The guanaco and llama have much
greater geographical ranges than the vicuña and alpaca,
which may have led to greater historical intraspecific
di¡erentiation, re£ected in greater diversity in nuclear
and mtDNA (where we found 21 guanaco haplotypes, as
opposed to five in the vicuña).
Analysis of greater
numbers of microsatellite loci (only four were used here)
is highly desirable, and could include some of those
described by Sarno et al. (2000), where at least two
additional loci suggest non-overlapping allele distributions
in the wild species. Archaeozoological evidence is
inconclusive concerning llama domestication, but it is
possible that there may have been more than one centre
(Wheeler 1995).
The suggestion of substantial mitochondrial introgression
in the alpaca and nuclear introgression in the
llama is substantiated when admixture is measured for
both marker types. The relatively low estimated admixture
proportion of vicu·a mtDNA present in the alpaca
(0.31) is in contrast with the high proportion estimated
for the microsatellites (0.82-0.90). Further, the extremely
low admixture proportion of vicu·a mtDNA in the llama
(0.02) contrasts strongly with microsatellite estimates
(0.22-0.39), which also suggest substantial nuclear admixture
in the llama. These data must, however, be treated
with caution, since the assumptions underlying the
admixture models used in this analysis are highly likely to
be violated in South American camelids, where introgression
events are likely to have been recurrent, and
may have increased in frequency during and after the
Spanish conquest. Furthermore, the drastic reductions in
domestic South American camelid populations associated
with the conquest are likely to have had signi¢cant and
unpredictable impacts on allele frequencies.
Inspection of the distribution of the three-locus
genotypes in modern alpacas and llamas conforms many
of the above ¢ndings. Only 27% of alpacas are mitochondrially
`vicuña’, although 40% of alpaca possess only
vicu·a microsatellite alleles with guanaco mtDNA haplotypes.
Such a pattern suggests that introgression of
guanaco (or, more likely, llama) mtDNA some time in the
past may have occurred recurrently within alpaca populations,
but may have been accompanied more recently
by a reversion to line or stock breeding within local alpaca
populations.
The lack of written records means that any such inference
is speculative. Table 3 suggests that, in contrast,
mitochondrial introgression has occurred much less
frequently in the llama. However, although table 3
suggests that nuclear introgression is similar in the alpaca
and llama, the expanded microsatellite dataset suggests
that it has occurred at a higher level in the llama
(possibly two or three times higher than in the alpaca
from the admixture analysis), which may partly account
for the more dispersed factorial correspondence pattern;
this warrants further investigation.
The implications of these data are potentially important
for the way in which these genetic resources are
managed in the future. In our sample, only 35% of domestic animals have not undergone any detectable
hybridization based on mtDNA and the two diagnostic
microsatellites. In particular, there are a very large
number of detectable hybrids within the alpaca population
(80%), which are accentuated when using mtDNA.
Also, 40% of llama show detectable signs of hybridization,
but mitochondrial introgression is virtually absent.
During the last 20-25 years large-scale hybridization
between llamas and alpacas has been carried out in Peru
(Bustinza 1989).
Speci¢cally, male alpacas have been bred
to female llamas to increase the population of animals
producing higher priced `alpaca’ ¢bre, and male llamas
have been bred to female alpacas to obtain greater fleece
weights and, thus, increased income.With sale price traditionally
determined by weight, and no consideration given
to fineness, the quality of alpaca fibre has decreased markedly
over the past 25 years.
Indigenous Quechua- and
Aymara-speaking herders subdivide the hybrids into
llamawari or waritu (llama-like) and pacowari or wayki
(alpaca-like), respectively, depending upon physical
appearance (Flores Ochoa 1977; Dransart 1991b).
The
o¡spring are fertile, tend to be intermediate in size and
can be backcrossed to either parental type. Further, recent
intensive selection for white £eece in modern alpaca may
also have involved bidirectional hybridization. A combination
of these practices and our results could explain the
taxonomic confusion surrounding the domestic forms in
the recent past, as it is likely that many specimens used in
previous taxonomic studies were hybrids.
Given the extreme hybridization in present-day
alpacas, DNA analysis has been critical in resolving the
origin of this domestic form. Since our results suggest the
vicu·a as the ancestor of the alpaca, we propose that
the classi¢cation of the alpaca should be changed from
Lama pacos L. to Vicugna pacos L.
We suggest, therefore, that the degeneration of quality
and value in present-day alpacas and llamas has been due
to extensive hybridization, probably beginning with the
conquest and continuing to the present day.While it was
believed that these crosses were between di¡erent forms
of a single domestic animal descended from the guanaco,
there was little concern about the economic impact of
such introgression.
However, given that the alpaca is
probably descended from the vicu·a, the negative impact
of such crosses is now evident. The use of DNA analysis to
identify and eliminate hybrid animals from the breeding
pool is essential, since the antiquity of the ongoing hybridization
process makes it impossible to accurately identify
all hybrids on the basis of phenotypic characteristics.
Additionally, the knowledge that the alpaca is descended
from the vicuña opens new routes for the improvement of
alpaca fibre production, not only through the identification
of hybrids and their elimination from pure-bred elite
herds, but also via the backcrossing of pure-bred alpacas
to their vicuña ancestor in order, possibly, to improvefibre fineness.
While 90% of the alpaca ¢bre produced in Peru today
has a diameter of greater than 25 mm and fetches low
prices on the world market (US$3-30per kg, 1980-1995),
pre-conquest animals produced fibre of 17-22mm in
diameter (Wheeler et al. 1995), similar to cashmere (15-
17 mm; US$60-120 per kg, 1980-1995). It is possible,
therefore, that identification of the remaining pure alpacas may aid in recovery of the fine fibre characteristics
of pre-conquest animals.
The knowledge that the alpaca is the domestic vicuña
also necessitates a re-evaluation of vicuña conservation
policy. Although the vicuña has been listed as endangered
under the Convention on International Trade in
Endangered Species of Wild Fauna and Flora (CITES
Appendix I) since its inception in 1975, all Peruvian
vicuñas and large segments of the Chilean, Argentine
and Bolivian populations have been reclassi¢ed as threatened
(CITES Appendix II), permitting controlled
commercialization of live-shorn fibre.
With unprocessedfibre currently valued at ca. US$405 per kg, vicuña fleece
is the most expensive natural fibre in the world, and
represents an important potential source of income for
the extremely poor rural populations on whose lands the
animals live. To date, Peru’s rational use policy has
produced an important increase in vicuña numbers, but
demands for greater control over the species through
construction of fences, intensive rearing and selection are
growing.
Judging by the alpaca, such interventions will,
in the long run, lead to a deterioration of fibre quality
and fi¢neness (which, at 12-14 mm, is the basis of its value),
and increased limitation on movement, especially of the
non-territorial male bachelor bands, represents a significant new threat to this species (Wheeler et al. 2001).
We acknowledge the following people who generously helped by
providing information, samples or permits for this study. In
Argentina: M. Knobel, Argentine Guanaco Products, Esquel; S.
Poncet; Eduardo Frank, Universidad de Cordoba; Freddy Sossa,
EU Supreme Project, Jujuy; Gustavo Rebu¤, INTA Abra
Pampa; Daniel Almeida, INTA Bariloche; Hector Guillermo
Villanueva, Recursos Naturales, Salta; Teresa Raquel Chalabe,
Universidad de Salta; G. Moseley, BBSRC^IGER, Aberystwyth,
UK. In Chile: Hernan Torres, Conservation International,
Washington DC, USA; Eduardo Nu·ez and Rafael Fernández,
CONAF, Arica; Calogero Santoro, Universidad de Tarapacá,
Arica. In Peru: Alfonso Martinez, Domingo Hoces, Jorge Herrera
and Marco Antonio Zuñiga, CONACS, Lima; Alex
Montufar and Marco Antonio Escobar, CONACS, Puno;
Roberto Bombilla, CONACS, Junín; Carlos Ponce del Prado,
Conservación Internacional, Lima; Maximo Gamarra and Santiago
Baudilio, SAIS Tupac Amaru, Pachacayo; Rosa Perales,
Jose Alva and Néstor Falcón, Facultad de Medicina Veterinaria,
Universidad Nacional Mayor de San Marcos, Lima; Leoncio
Ruiz Ríos, Congreso Constituyente del Perí; Clive Woodham,
Labvetsur, Arequipa; Felipe San Martín, Instituto Veterinario
de Investigaciones Tropicales y de Altura, Lima. Nicola
Anthony, Peter Arctander, Mark Beaumont, Jon Bridle, Kate
Byrne, Loune© s Chikhi, Gordon Luikart, Benoit Goossens and
two anonymous referees provided invaluable comments on the
manuscript, and Georgio Bertorelle gave valuable advice on the
admixture analysis. This study was supported by the Institute of
Zoology, InstitutoVeterinario de InvestigacionesTropicales y de
Altura, Natural Environment Research Council grant GST/02/828 to H.F.S. and J.C.W. and Darwin Initiative grant 162/06/126 to H.F.S, J.C.W and M.W.B.
REFERENCES
Bahn, P. G. 1994 Archaeozoologyötime for a change. Nature
367, 511-512.
Bandelt, H. J., Forster, P. & RÎhl, A. 1999 Median joining
networks for inferring intraspeci¢c phylogenies. Mol. Biol.
Evol. 16, 37-48
Belkhir, L. 1999 GENETIX v. 4.0. Belkhir Biosoft, Laboratoire des
Genomes et Populations, Université Montpellier II, France.
Benzécri, J. P. 1973 L’analyse des donnees:T. 2, I’analyse des correspondances.
Paris: Dunod.
Bertorelle, G. 1998 ADMIX 1_0. See www.fe.it/genetica/Giorgio/
giorgio_soft.html#ADMIX.
Bertorelle, G. & Exco¤er, L. 1998 Inferring admixture proportions
from molecular data.Mol. Biol. Evol. 15, 1298-1311.
Bradley, D. G., MacHugh, D. E., Cunningham, P., Loftus, R. T.
1996 Mitochondrial diversity and the origins of African and
European cattle. Proc. Natl Acad. Sci. USA 93, 5131-5135.
Bruford, M. W., Hanotte, O., Brook¢eld, J. F. Y. & Burke, T.
1998 Single and multilocus DNA ¢ngerprinting. In Molecular
genetic analysis of populations: a practical approach, 2nd edn (ed.
A. R. Hoelzel), pp. 287-336.Oxford University Press.
Bustinza,V. 1989 Algunas consecuencias de la agresión cultural
en la ganadería Andina. In Crianza de llamas y alpacas en los
Andes (ed. PAL/PRATEC), pp. 115^130. Lima, Talleres
Gra¢cos de ART Lautrec, S.R.L.
Chakraborty, R., Kamboh,M. I., Nwankwo,M. & Ferrell, R. E.
1992 Caucasian genes in American blacksönew data. Am. J.
Hum. Genet. 50, 145-155.
Dransart, P. 1991a Llamas, herders and the exploitation of raw
materials in the Atacama desert. World Archaeol. 22,
304-319.
Dransart, P. Z. 1991b Fibre to fabric: the role of fibre in camelid
economies in prehispanic and contemporary Chile. DPhil.
thesis, University of Oxford, UK.
Exco¤er, L. 1993 MINSPNET. See http://lgb.unige.ch/software/
win/min-span-net/.
Flores Ochoa, J. A. (ed.) 1977 Pastores de alpacas de los Andes.
In Pastores de puna, pp.15-52. Lima: Instituto de Estudios
Peruanos. Goldstein, D. B., Linares, A. R., Cavalli-Sforza, L. L. &
Feldman, M. W. 1995 An evaluation of genetic distances for
use with microsatellite loci. Genetics 139, 463-471.
Hall, S. J. G. & Bradley, D. G. 1995 Conserving livestock breed
diversity.Trends Ecol. Evol. 10, 267-270.
Hiendleder, S., Mainz, K., Plante, Y. & Lewalski, H. 1998
Analysis of mitochondrial DNA indicates that domestic sheep
are derived from two ancestral maternal sources: no evidence
for contributions from urial and argali sheep. J. Hered. 89,
113-120.
Kruskal, J. B. 1956 On the shortest spanning subtree of the
graph and the travelling salesman problem. Proc. Am. Math.
Soc. 7, 48-57.
Lang, K. D. M.,Wang,Y. & Plante,Y. 1996 Fifteen polymorphic
dinucleotide microsatellites in llamas and alpacas. Anim.
Genet. 27, 293.
Luikart, G., Gielly, L., Exco¤er, L., Vigne, J. D., Bouvet, J. &
Taberlet, P. 2001Multiple maternal origins and weak phylogeographic
structure in domestic goats. Proc. Natl Acad. Sci.
USA 98, 5927-5932.
MacHugh, D. E., Shriver, M. D., Loftus, R.T., Cunningham, P.& Bradley, D. G. 1997 Microsatellite DNA variation and the
evolution, domestication and phylogeography of taurine and
zebu cattle (Bos taurus and Bos indicus). Genetics 146, 1071-1086.
Minch, E. 1999 MICROSAT v. 1.5. See http://hpgl.stanford.edu/projects/microsat/.
Penedo, M. C. T., Casetano, A. R. & Cordova, K. I. 1998
Microsatellite markers for South American camelids. Anim.
Genet. 29, 411-412.
Reigadas, M. C. 1993 La punta del ovillo: determinación de
domesticación y pastoreo a partir del análisis microscopico de fibras y foliculos pilosos de camélidos. Arqueologica 2, 9-52.
Reynolds, J., Weir, B. S. & Cockerham, C. C. 1983 Estimation
of the co-ancestry coeficient-basis for a short-term genetic
distance. Genetics 105, 767-779.
Sarno, R. J., David, V. A., Franklin, W. L., O’Brien, S. J. &
Johnson, W. E. 2000 Development of microsatellite markers
in the guanaco Lama guanicoe: utility for South American
camelids. Mol. Ecol. 9, 1922-1924.
Stanley, H. F., Kadwell, M. & Wheeler, J. C. 1994 Molecular
evolution of the family Camelidae: a mitochondrial study.
Proc. R. Soc. Lond. B 256, 1-6.
Vila, C., Leonard, J. A., Gotherstrom, A., Marklund, S.,
Sandberg, K., Liden, K.,Wayne, R. K. & Ellegren, H. 2001
Widespread origins of domestic horse lineages. Science 291,
474-477.
Wheeler, J. C. 1995 Evolution and present situation of the South
American Camelidae. Biol. J. Linn. Soc. 54, 271-295.
Wheeler, J. C., Russel, A. J. F. & Redden, H. 1995 Llamas and
alpacas: pre-conquest breeds and post-conquest hybrids. J.
Archaeol. Sci. 22, 833-840.
Wheeler, J. C., Fernandez, M., Rosadio, R., Hoces, D.,
Kadwell, M. & Bruford, M. W. 2001 Diversidad genetica y
Manejo de Poblaciones de Vicuñas en el Peru. RIVEP
1(Suppl.), 170-183.
As this paper exceeds the maximum length normally permitted,
the authors have agreed to contribute to production costs.
|

